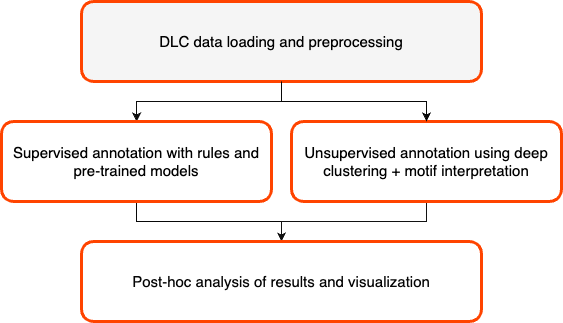
Welcome to DeepOF!
A suite for postprocessing time-series extracted from videos of freely moving rodents using DeepLabCut and SLEAP.

Getting started
You can use this package to either extract pre-defined motifs from the time series (such as time-in-zone, climbing, basic social interactions) or to embed your data into a sequence-aware latent space to extract meaningful motifs in an unsupervised way! Both of these can be used within the package, for example, to automatically compare user-defined experimental groups. The package is compatible with single and multi-animal DLC 2.X, and SLEAP projects.
Installation
The easiest way to install DeepOF is to use pip. Create and activate a virtual environment with Python >=3.9 and <3.11, for example using conda:
conda create -n deepof python=3.9
Then, activate the environment and install DeepOF:
conda activate deepof
pip install deepof
Alternatively, you can download our pre-built Docker image, which contains all compatible dependencies:
# download the latest available image
docker pull lucasmiranda42/deepof:latest
# run the image in interactive mode, enabling you to open python and import deepof
docker run -it lucasmiranda42/deepof
Or use poetry:
# after installing poetry and cloning the DeepOF repository, just run
poetry install # from the main directory
NOTE: installation via pip is at the moment not compatible with Apple Silicon. If you’d like to install DeepOF on such machines, please use either poetry or Docker. You should also install hdf5 using homebrew, as described in this issue.
What you need
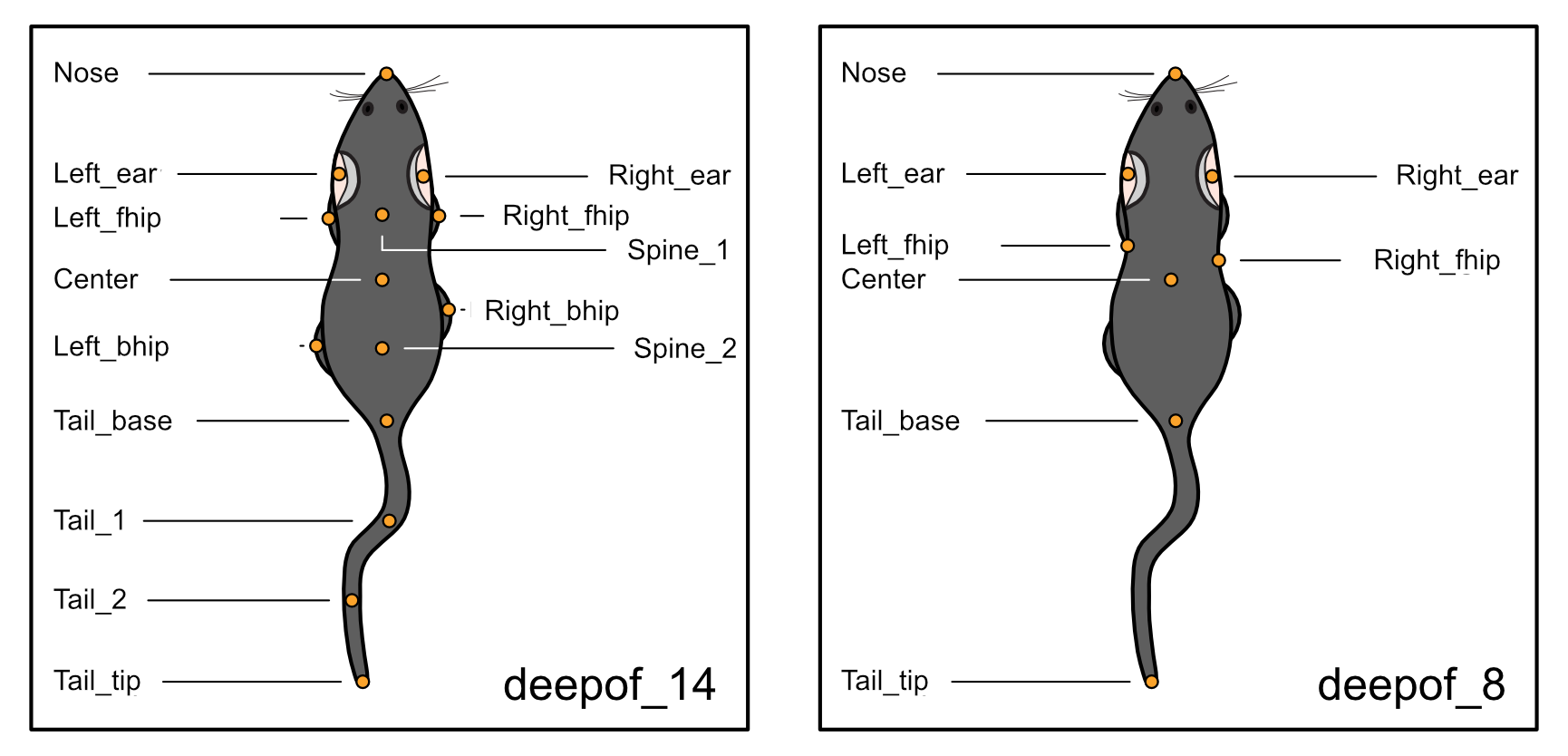
DeepOF relies heavily on DeepLabCut and SLEAP output. Thorough tutorials on how to get started with pose estimation using DLC can be found here, and for SLEAP here. Once your videos are processed and tagged, you can use DeepOF to extract and annotate your motion-tracking time-series. While many features in DeepOF can work regardless of the set of labels used, we currently recommend using videos from a top-down perspective, and follow our recommended set of labels (which can be found in the full documentation page). Pre-trained models following this scheme, and capable of recognizing either C57Bl6 mice alone, or C57Bl6 and CD1 mice can be downloaded from our repository.
Basic usage
The main module with which you’ll interact is called `deepof.data`. Let’s import it and create a project:
import deepof.data
my_deepof_project = deepof.data.Project(
project_path=".", # Path where to create project files
video_path="/path/to/videos", # Path to DLC tracked videos
table_path="/path/to/tables", # Path to DLC output
project_name="my_deepof_project", # Name of the current project
exp_conditions={exp_ID: exp_condition} # Dictionary containing one or more experimental conditions per provided video
bodypart_graph="deepof_14" # Labelling scheme to use. See the last tutorial for details
)
This command will create a `deepof.data.Project` object storing all the necessary information to start. There are
many parameters that we can set here, but let’s stick to the basics for now.
One you have this, you can run you project using the `.create()` method, which will do quite a lot of computing under
the hood (load your data, smooth your trajectories, compute distances, angles, and areas between body parts, and save all
results to disk). The returned object belongs to the `deepof.data.Coordinates` class.
my_project = my_project.create(verbose=True)
Once you have this, you can do several things! But let’s first explore how the results of those computations mentioned are stored. To extract trajectories, distances, angles and/or areas, you can respectively type:
my_project_coords = my_project.get_coords(center="Center", polar=False, align="Nose", speed=0)
my_project_dists = my_project.get_distances(speed=0)
my_project_angles = my_project.get_angles(speed=0)
my_project_areas = my_project.get_areas(speed=0)
Here, the data are stored as `deepof.data.table_dict` instances. These are very similar to python dictionaries
with experiment IDs as keys and pandas.DataFrame objects as values, with a few extra methods for convenience. Peeping
into the parameters you see in the code block above, `center` centers your data (it can be either a boolean or
one of the body parts in your model! in which case the coordinate origin will be fixed to the position of that point);
`polar` makes the `.get_coords()` method return polar instead of Cartesian coordinates, and `speed`
indicates the derivation level to apply (0 is position-based, 1 speed, 2 acceleration, 3 jerk, etc). Regarding
`align` and `align-inplace`, they take care of aligning the animal position to the y Cartesian axis: if we
center the data to “Center” and set `align="Nose", align_inplace=True`, all frames in the video will be aligned in a
way that will keep the Center-Nose axis fixed. This is useful to constrain the set of movements that one can extract
with our unsupervised methods.
As mentioned above, the two main analyses that you can run are supervised and unsupervised. They are executed by
the `.supervised_annotation()` method, and the `.deep_unsupervised_embedding()` methods of the `deepof.data.Coordinates`
class, respectively.
supervised_annot = my_project.supervised_annotation()
gmvae_embedding = my_project.deep_unsupervised_embedding()
The former returns a `deepof.data.TableDict` object, with a pandas.DataFrame per experiment containing a series of
annotations. The latter is a bit more complicated: it returns a series of objects that depend on the model selected (we
offer three flavours of deep clustering models), and allow for further analysis comparing cluster expression and dynamics.
That’s it for this (very basic) introduction. Check out the tutorials too see both pipelines in action, and the full API reference for details!
Tutorials
Formatting your data
Supervised and unsupervised pipelines
Advanced usage
Cite us!
If you use DeepOF in your research, please consider citing:
Contributing and support
If you’d like to contribute to DeepOF, please check out our contributing guidelines. If you’d like to report a bug, suggest a new feature, or need help with general usage, please open an issue in our issue tracker.


