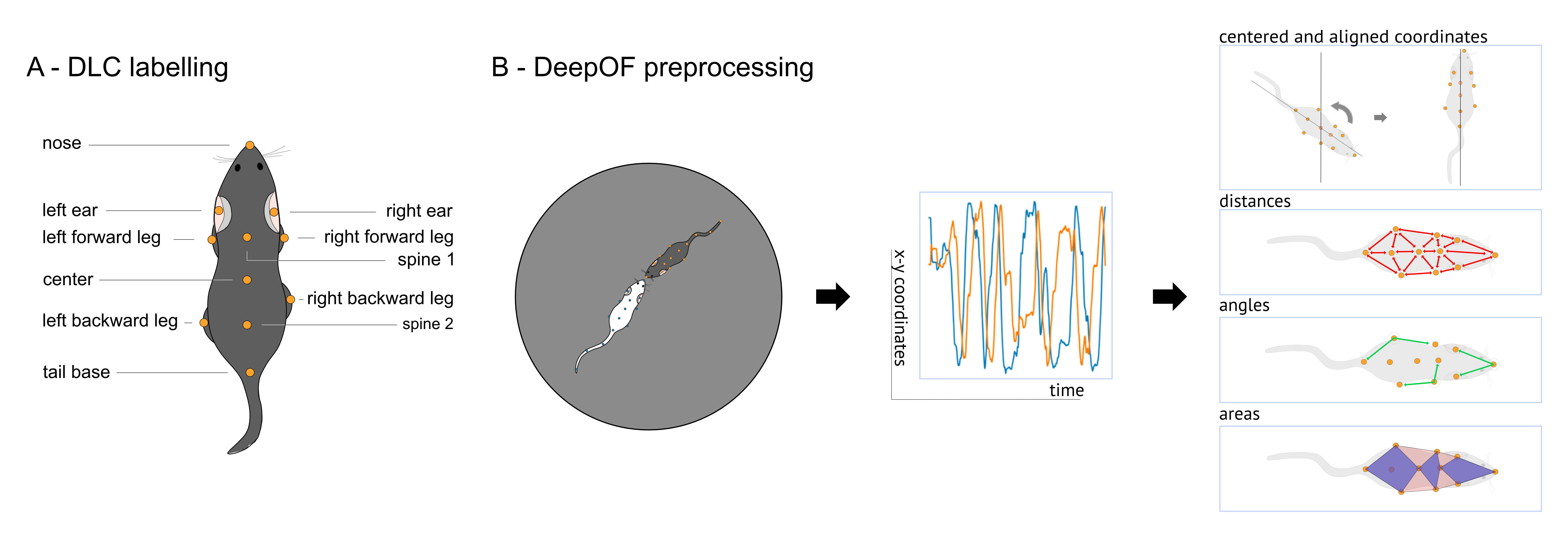
Formatting your data: feature extraction from motion tracking output
What we’ll cover:
Create and run a project.
Load a previously generated project.
Interact with your project: generate coordinates, distances, angles, and areas.
Exploratory visualizations: heatmaps and processed animations.
[1]:
# # If using Google colab, uncomment and run this cell and the one below to set up the environment
# # Note: becasuse of how colab handles the installation of local packages, this cell will kill your runtime.
# # This is not an error! Just continue with the cells below.
# import os
# !git clone -q https://github.com/mlfpm/deepof.git
# !pip install -q -e deepof --progress-bar off
# os.chdir("deepof")
# !curl --output tutorial_files.zip https://datashare.mpcdf.mpg.de/s/knF7t78isQuIAr0/download
# !unzip tutorial_files.zip
# os.kill(os.getpid(), 9)
[2]:
# os.chdir("deepof")
# import os, warnings
# warnings.filterwarnings('ignore')
Let’s start by importing some packages. We’ll use python’s os library to handle paths, pickle to load saved objects, pandas to load data frames, and the data entry API within DeepOF, located in deepof.data
[3]:
import os
import pandas as pd
import pickle
import deepof.data
We’ll also need some plotting gear:
[4]:
import deepof.visuals
import matplotlib.pyplot as plt
import seaborn as sns
Creating and running a project
With that out of the way, the first thing to do when starting a DeepOF project is to load your videos and DeepLabCut tracking files into a deepof.data.Project object.
Like depicted in the cell below, the three crucial parameters to input are the project, video and tab paths.
project_path specifies where the project folder containing all processing and output files will be created.
video_path points towards where your DLC or SLEAP labelled videos are stored.
similarly, table_path should point to the directory containing all tracking files (see section on supported input files below).
last but not least, you can give your project a name (optional; deepof_project by default).
The dataset used in this tutorial is a subset of the social interaction (SI) dataset used in DeepOF’s original paper. It contains six 10-minute-long videos with two animals (a C57Bl6 and a CD1) tracked in a round arena, tracked with DeepLabCut. Three of the C57Bl6 mice have been exposed to chronic social defeat stress (CSDS).
[5]:
my_deepof_project_raw = deepof.data.Project(
project_path=os.path.join("tutorial_files"),
video_path=os.path.join("tutorial_files/Videos/"),
table_path=os.path.join("tutorial_files/Tables/"),
project_name="deepof_tutorial_project",
arena="circular-autodetect",
animal_ids=["B", "W"],
table_format="h5",
video_format=".mp4",
exclude_bodyparts=["Tail_1", "Tail_2", "Tail_tip"],
video_scale=380,
smooth_alpha=1,
exp_conditions=None,
)
As you may see, there are some extra (optional) parameters in the call above. These specify some details on how the videos and tracks will be processed. Some include:
arena: some functions within DeepOF (as you will see in the next tutorial on supervised annotations) require the program to detect the arena. The following section explains how to handle this step in more detail.
animal_ids: in case more than one animal is present in the dataset, the IDs assigned to each during tracking need to be specified as a list. For SLEAP projects, this will be detected automatically, but can be overriten.
exclude_body_parts: while DeepOF originally relies on 14 body part tracking models, body parts along the tail are no longer used. This step could be bypassed using smaller models following an 11-body-part scheme, such as the one presented in the landing page of the documentation. Custom labelling schemes are also supported (see tutorials).
video_scale: diameter of the arena (in mm) if circular. In the case of polygonal arenas, the length (in mm) of the first edge as specified in the GUI (see next section) should be provided.
smooth_alpha: smoothing intensity. The higher the value, the more smoothing is applied.
exp_conditions: dictionary with a video IDs as keys, and data frames with all experimental conditions as values. DeepOF will use this information to enable all sorts of comparisons between conditions, as we will see in the following tutorials. We’ll leave it blank for now and update it afterwards.
For more details, feel free to visit the full API reference.
Let’s then create our first project, by running the .create() method in our newly created object:
[6]:
my_deepof_project = my_deepof_project_raw.create(force=True)
Setting up project directories...
Loading trajectories...
Smoothing trajectories...
Interpolating outliers...
Iterative imputation of ocluded bodyparts...
Detecting arena...
Computing distances...
Computing angles...
Computing areas...
Done!
NOTE: the force parameter allows DeepOF to override an existing project in the same directory. Use with caution!
NOTE 2: the cell above can take a significant amount of time to run in Google colab. Feel free to skip and continue with the loaded results below.
As you will see, this organizes all required file into a folder in the specified path. Moreover, some processing steps are computed on the go, such as distances, angles and areas. This makes it easier for DeepOF to load all required features later on, without the need to compute them every time.
Arena detection
As mentioned above, one of the key aspects of project creation involves setting up the arena detection mechanism, which will be used in downstream tasks, such as climbing detection and overall video scaling.
You can contrAlong these lines, the package provides tools for detecting elliptical arenas specifically (as seen in this tutorial), and general polygonal shapes (such as squares, rectangles, Y-mazes, etc).
In principle, DeepOF can detect your arenas automatically by relying in SAM (Segment Anything Model), a state-of-the-art image segmentation deep learning model. By selecting arena='circular-autodetect', or arena='polygonal-autodetect', users can benefit from this approach. A folder named Arena_detection will be created in your project directory, which contains samples of the detected arenas for all videos, so you can visually inspect if DeepOF did
a good job.
Moreover, manual annotation can be selected using arena='circular-manual', and arena='polygonal-manual'. In this case, you will see a window per video appear, allowing you to manually mark where the arena is with just a few clicks. All results will be stored for further processing.
Here, clicking anywhere in the video will make an orange marker appear. In the case of polygonal arenas, at least a marker per corner should be used. When dealing with circular (or elliptical) arenas, DeepOF will fit an ellipse to the marked points after a minimum of 5 clicks. The ellipse can always be refined by adding more markers. Once finished with a video, press q to save and move to the next one. If you made a mistake and would like to correct it, press d to delete the last added
marker. Moreover, after you tagged at least one video, you’ll see the p option appear, which will copy the last marked arena to all remaining videos in the dataset.
NOTE: If you select arena='polygonal-autodetect', you will still be prompted with the GUI to select the arena just once, so SAM roughly knows how the segmentation it should return looks like. Detection in all remaining videos will be automatic. This is not the case for arena='circular-autodetect', as we already know we’re looking for a circle.
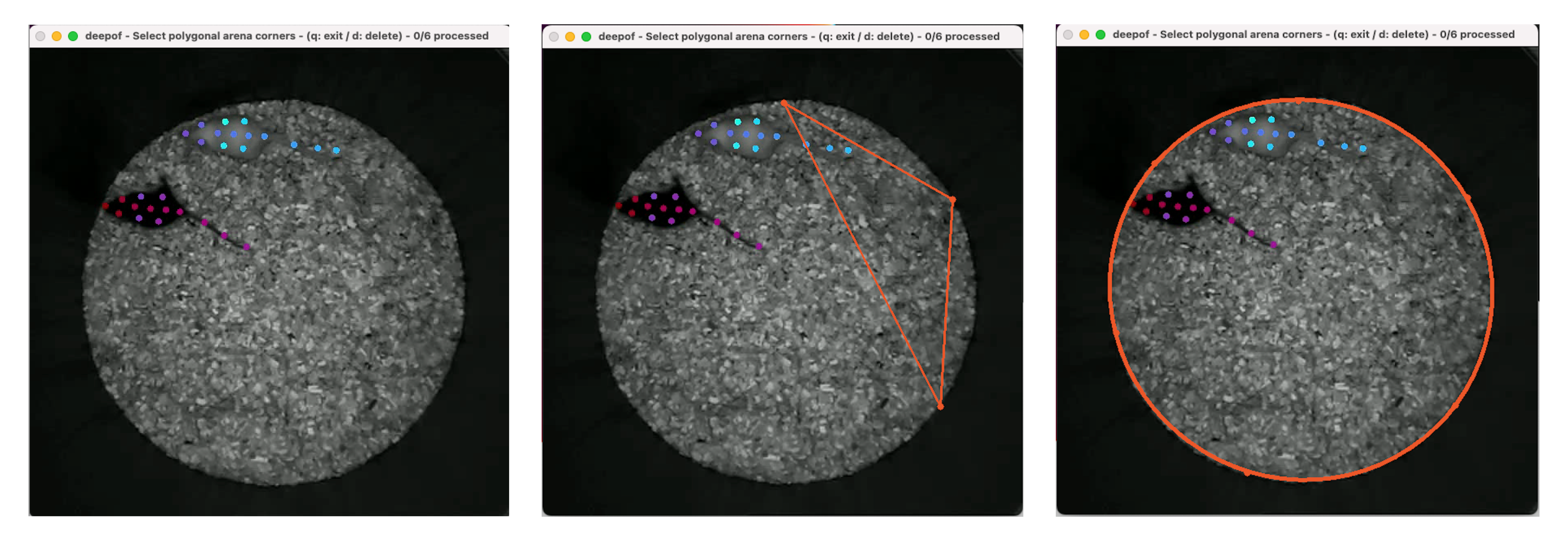
One last thing is that, in polygonal arenas, the first edge you input will be used to scale the coordinates to the proper distances (pixels to millimeters). Make sure you always mark the same edge first, and that it coincides with the length passed to the “video_scale” parameter when creating the project. In saved images of automatically detected polygons, you’ll see orange circles marking two arena corners. These should match the first edge you selected when prompting SAM, ensuring data is properly scaled.
If after all this work you make a mistake while labelling, or see that automatic detection failed in some cases, don’t worry! You can always edit manually annotated arenas (or rerun automatic annotation) for specific videos using the .edit_arenas() method. Just pass a list with the IDs of the videos you would like to relabel, and the GUI will pop up once again. The same methods described above can be passed to the arena_type parameter.
NOTE: If you don’t pass any video IDs, all videos will be selected by default.
[7]:
my_deepof_project.edit_arenas(
videos=['20191204_Day2_SI_JB08_Test_54', '20191204_Day2_SI_JB08_Test_62'],
arena_type="circular-autodetect",
)
Editing 2 arenas
Done!
Supported input tracking files:
DeepOF currently supports input from both DeepLabCut and SLEAP. While DeepOF will try to detect automatically which file type you’re trying to use, this can be forced with the table_format parameter in the deepof.data.Project() call depicted in the previous section.
For DeepLabCut, we support:
Single and multi-animal project CSV files (indicated simply with
table_format='csv'.Single and multi-animal project h5 files (indicated simply with
table_format='h5'.
For SLEAP, you can use:
SLP files (indicated as with
table_format='slp'.Raw .npy files (indicated with
table_format='npy'.h5 files (indicated with
table_format=analysis.h5.
Downstream processing should be identical regardless of the files selected, as DeepOF internally brings all these inputs into an equivalent representation. Let’s continue!
Loading a previously generated project
Once you ran your project at least once, it can be loaded without much effort from the path you specified in the first place (plus the project name -deepof_project, if you didn’t specify otherwise-).
[8]:
# Load a previously saved project
my_deepof_project = deepof.data.load_project("./tutorial_files/deepof_tutorial_project/")
Extend a previously generated project
If you’d like to add data (videos and tracks) to a previously processed project, you can use the extend method instead of create. Just pass the path to your previously processed project, and DeepOF will take care of merging the two. A new directory will be created with all files corresponding to the merged projects.
NOTE: Your old files will not be deleted.
[9]:
# Extend a previously saved project
my_deepof_project_raw.extend("./tutorial_files/deepof_tutorial_project/")
Loading previous project...
Processing data from 0 experiments...
No new experiments to process. Exiting...
Interacting with your project: generating coordinates, distances, angles, and areas.
That’s it for basic project creation. We now have a DeepOF project up and running! Before ending the tutorial, however, let’s explore the object that the commands above produced.
For starters, if we print it, we see it’s a DeepOF analysis of 6 videos. Furthermore, the object belongs to a custom class within DeepOF, called Coordinates. This class allows the package to store all sorts of relevant information required for further processing, as we’ll see below:
[11]:
print(my_deepof_project)
print(type(my_deepof_project))
deepof analysis of 6 videos
<class 'deepof.data.Coordinates'>
As described before, the .create() method runs most of the heavy preprocessing already, which allows us to extract features including coordinates, distances, angles, and areas. Let’s see how that works!
With the .get_coords() method, for example, we can obtain the processed (smooth and imputed) tracks for all videos in a dictionary. The returned objects are called table dictionaries (TableDict is the name of the class). They follow a dictionary-like structure, where each value is a data frame. They also provide a plethora of extra methods, some of which we’ll cover in these tutorials. Let’s retrieve these for one of the animals:
[12]:
my_deepof_project.get_coords(polar=False, center="Center", align="Spine_1")['20191204_Day2_SI_JB08_Test_54']
[12]:
| B_Spine_1 | B_Center | B_Left_bhip | B_Left_ear | B_Left_fhip | ... | W_Right_bhip | W_Right_ear | W_Right_fhip | W_Spine_2 | W_Tail_base | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | x | y | x | y | x | y | x | y | ... | x | y | x | y | x | y | x | y | x | y | |
| 00:00:00 | 0.0 | 17.041283 | 0.0 | 0.0 | 16.235626 | -9.286406 | 2.338149 | 35.826017 | 14.520708 | 16.191088 | ... | -11.115287 | -13.877286 | -11.056773 | 52.165913 | -10.837443 | 11.900607 | 1.303025 | -19.189055 | -9.987794 | -46.786090 |
| 00:00:00.039935995 | 0.0 | 16.475167 | 0.0 | 0.0 | 15.143549 | -10.150172 | 0.671672 | 35.836244 | 14.183559 | 14.703795 | ... | -13.884385 | -18.083860 | -11.634608 | 53.793769 | -13.457341 | 12.356739 | 1.828954 | -24.439299 | -1.620732 | -48.755033 |
| 00:00:00.079871991 | 0.0 | 16.446049 | 0.0 | 0.0 | 11.477393 | -12.396184 | 0.097058 | 35.005123 | 13.716176 | 13.898245 | ... | -15.550991 | -13.873025 | -12.439161 | 53.259569 | -14.284002 | 12.808460 | 0.793345 | -21.164099 | 1.765989 | -41.624561 |
| 00:00:00.119807987 | 0.0 | 15.217471 | 0.0 | 0.0 | 10.208930 | -14.160422 | -0.358944 | 35.315238 | 15.230370 | 11.435026 | ... | -14.469009 | -16.138564 | -11.961743 | 52.265874 | -14.571504 | 13.210702 | 1.309584 | -22.165508 | 2.815459 | -42.444154 |
| 00:00:00.159743982 | 0.0 | 15.037661 | 0.0 | 0.0 | 10.836279 | -15.220016 | 0.861588 | 34.324995 | 14.984768 | 10.471671 | ... | -14.028915 | -18.169221 | -19.961073 | 49.485545 | -14.900604 | 11.323389 | 1.955114 | -22.447853 | 2.289354 | -44.418900 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800320021 | 0.0 | 17.383252 | 0.0 | 0.0 | 15.964064 | -11.925356 | 10.656661 | 35.405103 | 14.810418 | 11.274649 | ... | -20.815692 | -6.861448 | -17.302919 | 31.105013 | -12.210518 | 10.521213 | -6.286187 | -15.291244 | -18.448989 | -26.719926 |
| 00:09:58.840256017 | 0.0 | 15.069470 | 0.0 | 0.0 | 15.782188 | -13.477302 | 7.241669 | 39.238723 | 14.817890 | 9.091584 | ... | -20.930728 | -7.043085 | -17.317452 | 31.011691 | -12.259446 | 10.426808 | -6.220324 | -15.480332 | -18.379192 | -26.895317 |
| 00:09:58.880192012 | 0.0 | 19.435457 | 0.0 | 0.0 | 12.847697 | -11.565332 | 7.622541 | 42.242592 | 14.851511 | 12.187561 | ... | -20.912197 | -7.052485 | -17.362287 | 30.948820 | -12.239039 | 10.390575 | -6.179682 | -15.503151 | -18.331690 | -26.956528 |
| 00:09:58.920128008 | 0.0 | 19.435457 | 0.0 | 0.0 | 12.847697 | -11.565332 | 7.622541 | 42.242592 | 14.851511 | 12.187561 | ... | -20.912197 | -7.052485 | -17.362287 | 30.948820 | -12.239039 | 10.390575 | -6.179682 | -15.503151 | -18.331690 | -26.956528 |
| 00:09:58.960064004 | 0.0 | 19.435457 | 0.0 | 0.0 | 12.847697 | -11.565332 | 7.622541 | 42.242592 | 14.851511 | 12.187561 | ... | -20.912197 | -7.052485 | -17.362287 | 30.948820 | -12.239039 | 10.390575 | -6.179682 | -15.503151 | -18.331690 | -26.956528 |
14999 rows × 44 columns
Note that there are a few parameters you can pass to the .get_coords() method. If “polar” is set to True, polar coordinates will be used, instead of Cartesian. Both “center” and “align” control how translational and rotational variance are removed from the data: the former will use the specified body part as [0, 0] (or the center of the arena, if set to “arena”). The latter will rotate the mice on each time point, to align the line connecting the body parts specified in the “center” and
“align” parameters with the y-axis.
Furthermore, not only processed coordinates can be retrieved, but also distances, angles, and areas with the .get_distances(), .get_angles(), and .get_areas(), respectively.
[13]:
my_deepof_project.get_distances()['20191204_Day2_SI_JB08_Test_54']
[13]:
| (B_Nose, B_Right_ear) | (B_Tail_base, W_Tail_base) | (W_Center, W_Right_fhip) | (B_Center, B_Left_fhip) | (W_Left_ear, W_Spine_1) | (W_Left_ear, W_Nose) | (B_Tail_base, W_Nose) | (W_Center, W_Spine_2) | (B_Right_bhip, B_Spine_2) | (W_Center, W_Spine_1) | ... | (B_Right_ear, B_Spine_1) | (W_Spine_2, W_Tail_base) | (B_Left_ear, B_Nose) | (B_Center, B_Spine_1) | (B_Nose, W_Tail_base) | (W_Nose, W_Right_ear) | (B_Left_bhip, B_Spine_2) | (W_Center, W_Left_fhip) | (B_Nose, W_Nose) | (B_Spine_2, B_Tail_base) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00:00:00 | 18.781728 | 97.709786 | 13.041369 | 17.621480 | 27.055962 | 18.834943 | 148.048046 | 15.583439 | 11.304016 | 19.977755 | ... | 14.239388 | 24.159109 | 15.890697 | 13.807436 | 115.086175 | 22.834626 | 15.229008 | 18.574999 | 197.209343 | 14.568245 |
| 00:00:00.039935995 | 16.395499 | 102.888817 | 14.802899 | 16.552894 | 25.757160 | 19.117538 | 156.962058 | 19.856936 | 11.571620 | 21.885587 | ... | 14.693174 | 19.898728 | 15.271448 | 13.348749 | 118.018418 | 21.852281 | 15.199081 | 20.296875 | 201.897777 | 14.454055 |
| 00:00:00.079871991 | 15.057738 | 112.513259 | 15.544880 | 15.821263 | 27.572622 | 22.222081 | 165.295346 | 17.159928 | 11.412604 | 19.920155 | ... | 14.429184 | 16.596494 | 15.170325 | 13.325157 | 122.652716 | 24.791305 | 14.600725 | 20.121680 | 202.543917 | 14.753496 |
| 00:00:00.119807987 | 15.112614 | 119.626219 | 15.936129 | 15.431171 | 29.026338 | 20.982981 | 171.180259 | 17.990578 | 11.977952 | 20.211183 | ... | 15.581831 | 16.475699 | 16.038286 | 12.329721 | 122.474901 | 24.705419 | 14.741844 | 19.739289 | 201.439639 | 15.937254 |
| 00:00:00.159743982 | 16.235095 | 123.351461 | 15.163451 | 14.811991 | 26.424186 | 21.167892 | 174.253493 | 18.256880 | 12.857731 | 21.018731 | ... | 15.989769 | 17.803761 | 18.028338 | 12.184032 | 121.775639 | 22.790681 | 14.977541 | 20.680558 | 200.696460 | 15.523106 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800320021 | 22.674736 | 231.732647 | 13.059429 | 15.081384 | 18.866566 | 23.477526 | 172.650733 | 13.395564 | 13.647533 | 15.129049 | ... | 16.974596 | 13.522631 | 20.974634 | 14.084511 | 191.389072 | 19.277812 | 13.789807 | 15.207663 | 148.312574 | 14.714033 |
| 00:09:58.840256017 | 22.789396 | 228.571928 | 13.039802 | 14.085660 | 18.910177 | 23.502572 | 169.636565 | 13.517401 | 13.324808 | 15.079827 | ... | 17.378502 | 13.512709 | 16.403318 | 12.209805 | 186.729656 | 19.322534 | 13.468828 | 15.197649 | 144.670506 | 14.576626 |
| 00:09:58.880192012 | 21.730112 | 226.395893 | 13.008192 | 15.566291 | 18.928975 | 23.463719 | 168.414230 | 13.522330 | 13.673167 | 15.108134 | ... | 17.947812 | 13.529974 | 18.592353 | 15.747278 | 186.008429 | 19.336323 | 13.742711 | 15.241505 | 146.851047 | 15.374270 |
| 00:09:58.920128008 | 21.730112 | 226.395893 | 13.008192 | 15.566291 | 18.928975 | 23.463719 | 168.414230 | 13.522330 | 13.673167 | 15.108134 | ... | 17.947812 | 13.529974 | 18.592353 | 15.747278 | 186.008429 | 19.336323 | 13.742711 | 15.241505 | 146.851047 | 15.374270 |
| 00:09:58.960064004 | 21.730112 | 226.395893 | 13.008192 | 15.566291 | 18.928975 | 23.463719 | 168.414230 | 13.522330 | 13.673167 | 15.108134 | ... | 17.947812 | 13.529974 | 18.592353 | 15.747278 | 186.008429 | 19.336323 | 13.742711 | 15.241505 | 146.851047 | 15.374270 |
14999 rows × 26 columns
[14]:
my_deepof_project.get_angles()['20191204_Day2_SI_JB08_Test_54']
[14]:
| (B_Left_ear, B_Nose, B_Right_ear) | (B_Nose, B_Left_ear, B_Spine_1) | (B_Right_fhip, B_Center, B_Spine_2) | (B_Right_fhip, B_Center, B_Left_fhip) | (B_Right_fhip, B_Center, B_Spine_1) | (B_Spine_2, B_Center, B_Left_fhip) | (B_Spine_2, B_Center, B_Spine_1) | (B_Left_fhip, B_Center, B_Spine_1) | (B_Left_bhip, B_Spine_2, B_Center) | (B_Left_bhip, B_Spine_2, B_Right_bhip) | ... | (W_Left_bhip, W_Spine_2, W_Center) | (W_Left_bhip, W_Spine_2, W_Right_bhip) | (W_Left_bhip, W_Spine_2, W_Tail_base) | (W_Center, W_Spine_2, W_Right_bhip) | (W_Center, W_Spine_2, W_Tail_base) | (W_Right_bhip, W_Spine_2, W_Tail_base) | (W_Nose, W_Right_ear, W_Spine_1) | (W_Left_ear, W_Spine_1, W_Center) | (W_Left_ear, W_Spine_1, W_Right_ear) | (W_Center, W_Spine_1, W_Right_ear) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00:00:00 | 1.222277 | 1.422657 | 1.805680 | 1.281616 | 1.342828 | 0.880634 | 1.020852 | 1.157763 | 1.110616 | 1.293108 | ... | 0.931157 | 0.848570 | 0.884871 | 0.865821 | 0.905761 | 0.866423 | 1.181972 | 1.145697 | 1.162453 | 1.018039 |
| 00:00:00.039935995 | 1.306489 | 1.247439 | 1.370046 | 1.299858 | 1.296478 | 1.502996 | 1.503997 | 1.052589 | 1.150996 | 1.200270 | ... | 0.994129 | 0.891450 | 0.966073 | 0.853879 | 0.919997 | 0.829069 | 1.212442 | 1.149331 | 1.114826 | 1.044152 |
| 00:00:00.079871991 | 1.282906 | 1.033643 | 1.190446 | 1.295514 | 1.433251 | 1.331724 | 1.469462 | 1.398862 | 1.548047 | 1.722389 | ... | 1.017974 | 0.977607 | 0.900188 | 0.915664 | 0.838244 | 0.740774 | 1.069299 | 1.090081 | 1.214856 | 1.133077 |
| 00:00:00.119807987 | 1.251990 | 1.454675 | 1.765511 | 1.277808 | 1.363800 | 0.915767 | 1.064082 | 1.182411 | 1.116587 | 1.309047 | ... | 0.968484 | 0.885802 | 0.910080 | 0.903568 | 0.931403 | 0.882201 | 1.213669 | 1.177122 | 1.188797 | 1.039895 |
| 00:00:00.159743982 | 1.330860 | 1.264571 | 1.356475 | 1.286804 | 1.286837 | 1.450061 | 1.454712 | 1.065928 | 1.106752 | 1.156228 | ... | 1.003958 | 0.907370 | 0.988059 | 0.879502 | 0.953687 | 0.854115 | 1.254271 | 1.189981 | 1.148939 | 1.079375 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800320021 | 1.249109 | 1.107626 | 0.859565 | 0.832034 | 0.888055 | 0.753215 | 0.805548 | 0.793002 | 0.664003 | 0.694227 | ... | 0.006698 | 0.026468 | 0.016655 | 0.036679 | 0.036456 | 0.152219 | 0.049799 | 0.096655 | 0.082662 | 0.071047 |
| 00:09:58.840256017 | 1.097980 | 0.939944 | 0.779166 | 0.870837 | 0.946506 | 0.793762 | 0.862571 | 0.836612 | 0.737746 | 0.738258 | ... | 0.104709 | 0.167571 | 0.107108 | 0.170584 | 0.116224 | 0.140526 | 0.030954 | 0.026903 | 0.110395 | 0.121503 |
| 00:09:58.880192012 | 1.062318 | 1.104193 | 0.861663 | 0.853209 | 0.904947 | 0.707809 | 0.759548 | 0.837450 | 0.711801 | 0.763568 | ... | 0.054504 | 0.049245 | 0.078203 | 0.145142 | 0.174100 | 0.247217 | 0.180157 | 0.128315 | 0.004496 | 0.090246 |
| 00:09:58.920128008 | 1.271455 | 1.135542 | 0.861166 | 0.833963 | 0.890707 | 0.749906 | 0.802551 | 0.806550 | 0.667206 | 0.697214 | ... | 0.003272 | 0.021836 | 0.011959 | 0.027468 | 0.026288 | 0.141577 | 0.061570 | 0.109516 | 0.093470 | 0.063343 |
| 00:09:58.960064004 | 1.136102 | 0.979934 | 0.781150 | 0.872829 | 0.948545 | 0.795213 | 0.864082 | 0.841856 | 0.735044 | 0.735689 | ... | 0.099354 | 0.163372 | 0.102508 | 0.162928 | 0.108771 | 0.133632 | 0.052552 | 0.041997 | 0.097895 | 0.103528 |
14999 rows × 36 columns
[15]:
my_deepof_project.get_areas()['20191204_Day2_SI_JB08_Test_54']
[15]:
| B_head_area | B_torso_area | B_back_area | B_full_area | W_head_area | W_torso_area | W_back_area | W_full_area | |
|---|---|---|---|---|---|---|---|---|
| 00:00:00 | 287.018462 | 467.472460 | 513.431000 | 1697.086099 | 588.316685 | 575.287843 | 557.837678 | 2402.446220 |
| 00:00:00.039935995 | 298.006089 | 461.593907 | 511.611552 | 1664.187297 | 532.941463 | 711.610814 | 747.171324 | 2612.577515 |
| 00:00:00.079871991 | 283.907493 | 436.994147 | 491.022208 | 1571.659017 | 615.224965 | 659.274363 | 696.773878 | 2680.819039 |
| 00:00:00.119807987 | 336.017364 | 459.434553 | 495.609970 | 1674.157290 | 664.955274 | 698.278394 | 709.587295 | 2725.074795 |
| 00:00:00.159743982 | 338.565821 | 456.785954 | 510.514167 | 1706.762126 | 589.501549 | 713.393985 | 706.975280 | 2732.936140 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800320021 | 364.598702 | 512.092068 | 562.957165 | 1882.932433 | 516.368794 | 466.566421 | 607.161201 | 1974.352930 |
| 00:09:58.840256017 | 342.771541 | 514.231920 | 543.299019 | 1832.083470 | 517.334943 | 468.582411 | 611.765902 | 1979.455000 |
| 00:09:58.880192012 | 388.332158 | 529.127829 | 558.340318 | 1889.399535 | 516.875687 | 468.401897 | 613.444346 | 1980.834529 |
| 00:09:58.920128008 | 388.332158 | 529.127829 | 558.340318 | 1889.399535 | 516.875687 | 468.401897 | 613.444346 | 1980.834529 |
| 00:09:58.960064004 | 388.332158 | 529.127829 | 558.340318 | 1889.399535 | 516.875687 | 468.401897 | 613.444346 | 1980.834529 |
14999 rows × 8 columns
Last but not least, features can be merged using the .merge() method, which can yield combinations of features if needed. For example, the code in the following cell creates an object with both coordinates and areas per time point:
[16]:
my_deepof_project.get_coords().merge(my_deepof_project.get_areas())['20191204_Day2_SI_JB08_Test_54']
[16]:
| (B_Center, x) | (B_Center, y) | (B_Left_bhip, x) | (B_Left_bhip, y) | (B_Left_ear, x) | (B_Left_ear, y) | (B_Left_fhip, x) | (B_Left_fhip, y) | (B_Nose, x) | (B_Nose, y) | ... | (W_Tail_base, x) | (W_Tail_base, y) | B_head_area | B_torso_area | B_back_area | B_full_area | W_head_area | W_torso_area | W_back_area | W_full_area | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00:00:00 | 223.193283 | 184.124115 | 222.160935 | 202.799423 | 191.852982 | 166.609666 | 201.709718 | 187.509186 | 188.298004 | 147.322081 | ... | 321.353759 | 97.605996 | 287.018462 | 467.472460 | 513.431000 | 1697.086099 | 588.316685 | 575.287843 | 557.837678 | 2402.446220 |
| 00:00:00.039935995 | 222.081848 | 185.302109 | 221.340759 | 203.517593 | 192.753159 | 164.698335 | 201.841401 | 188.076859 | 194.544312 | 145.935454 | ... | 332.379546 | 98.838112 | 298.006089 | 461.593907 | 511.611552 | 1664.187297 | 532.941463 | 711.610814 | 747.171324 | 2612.577515 |
| 00:00:00.079871991 | 220.547333 | 186.246719 | 219.854218 | 203.126160 | 197.765198 | 159.669617 | 201.092896 | 184.567551 | 205.235306 | 142.500976 | ... | 350.856689 | 101.147659 | 283.907493 | 436.994147 | 491.022208 | 1571.659017 | 615.224965 | 659.274363 | 696.773878 | 2680.819039 |
| 00:00:00.119807987 | 218.467209 | 184.700256 | 216.118439 | 201.998322 | 202.173355 | 153.366486 | 199.648239 | 181.772736 | 214.989639 | 138.281066 | ... | 361.710663 | 101.918587 | 336.017364 | 459.434553 | 495.609970 | 1674.157290 | 664.955274 | 698.278394 | 709.587295 | 2725.074795 |
| 00:00:00.159743982 | 219.402542 | 182.458069 | 214.480026 | 200.481461 | 206.756088 | 150.536057 | 201.725708 | 177.796661 | 221.262161 | 133.663879 | ... | 369.073914 | 106.446114 | 338.565821 | 456.785954 | 510.514167 | 1706.762126 | 589.501549 | 713.393985 | 706.975280 | 2732.936140 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800320021 | 108.366647 | 302.971861 | 127.398353 | 297.067658 | 106.577644 | 339.902678 | 118.557281 | 318.548034 | 91.342528 | 360.831938 | ... | 267.896393 | 517.758606 | 364.598702 | 512.092068 | 562.957165 | 1882.932433 | 516.368794 | 466.566421 | 607.161201 | 1974.352930 |
| 00:09:58.840256017 | 108.605596 | 310.532362 | 127.530082 | 302.013032 | 104.571034 | 350.229231 | 120.285217 | 323.409210 | 93.433890 | 367.135740 | ... | 267.855316 | 517.770020 | 342.771541 | 514.231920 | 543.299019 | 1832.083470 | 517.334943 | 468.582411 | 611.765902 | 1979.455000 |
| 00:09:58.880192012 | 108.844546 | 318.092862 | 124.715041 | 311.241029 | 102.564423 | 360.555784 | 119.021164 | 334.388275 | 86.603620 | 377.042506 | ... | 267.961975 | 517.801758 | 388.332158 | 529.127829 | 558.340318 | 1889.399535 | 516.875687 | 468.401897 | 613.444346 | 1980.834529 |
| 00:09:58.920128008 | 108.844546 | 318.092862 | 124.715041 | 311.241029 | 102.564423 | 360.555784 | 119.021164 | 334.388275 | 86.603620 | 377.042506 | ... | 267.961975 | 517.801758 | 388.332158 | 529.127829 | 558.340318 | 1889.399535 | 516.875687 | 468.401897 | 613.444346 | 1980.834529 |
| 00:09:58.960064004 | 108.844546 | 318.092862 | 124.715041 | 311.241029 | 102.564423 | 360.555784 | 119.021164 | 334.388275 | 86.603620 | 377.042506 | ... | 267.961975 | 517.801758 | 388.332158 | 529.127829 | 558.340318 | 1889.399535 | 516.875687 | 468.401897 | 613.444346 | 1980.834529 |
14999 rows × 52 columns
Loading experimental conditions
So far, DeepOF does not know to which condition each animal belongs. This can be either set up when creating the project (as described above) or specified afterward using the .load_exp_conditions() method. We just need to pass the path to a CSV file containing all conditions per animal as extra columns. The only hard requirement is that the first column should have the experiment IDs.
Here is an example:
[17]:
pd.read_csv("./tutorial_files/tutorial_project/Coordinates/tutorial_exp_conditions.csv", index_col=0)
[17]:
| experiment_id | CSDS | |
|---|---|---|
| 0 | 20191204_Day2_SI_JB08_Test_54 | Nonstressed |
| 1 | 20191204_Day2_SI_JB08_Test_56 | Stressed |
| 2 | 20191204_Day2_SI_JB08_Test_61 | Stressed |
| 3 | 20191204_Day2_SI_JB08_Test_62 | Stressed |
| 4 | 20191204_Day2_SI_JB08_Test_63 | Nonstressed |
| 5 | 20191204_Day2_SI_JB08_Test_64 | Nonstressed |
Great! Now that we understand how the CSV should be formatted, let’s then load it onto our project:
[18]:
my_deepof_project.load_exp_conditions("./tutorial_files/tutorial_project/Coordinates/tutorial_exp_conditions.csv")
And we’re done!. Let’s explore what’s in there with the .get_exp_conditions property:
[19]:
print(my_deepof_project.get_exp_conditions)
{'20191204_Day2_SI_JB08_Test_54': CSDS
0 Nonstressed, '20191204_Day2_SI_JB08_Test_56': CSDS
1 Stressed, '20191204_Day2_SI_JB08_Test_61': CSDS
2 Stressed, '20191204_Day2_SI_JB08_Test_62': CSDS
3 Stressed, '20191204_Day2_SI_JB08_Test_63': CSDS
4 Nonstressed, '20191204_Day2_SI_JB08_Test_64': CSDS
5 Nonstressed}
We can see that the property retrieves a dictionary with all animal experiments as keys, and data frames with conditions as values. Although in this case we only have the CSDS condition, which can take two values (“Nonstressed” and “Stressed”, with three animals each), adding more just requires us to add extra columns to the CSV file.
Filtering DeepOF objects
Now that experimental conditions were added, we’ll explore some filtering tools that DeepOF provides for table dictionary objects.
For starters, imagine you want to subset your data to only contain stressed animals. You can do that with the .filter_condition() method, which takes a dictionary as input with the experimental condition to filter on as key, and the value you’d like to keep as value:
[20]:
# Let's use coords as an example
coords = my_deepof_project.get_coords()
print("The original dataset has {} videos".format(len(coords)))
# Let's keep only those experiments where the subject is stressed:
coords = coords.filter_condition({"CSDS": "Stressed"})
print("The filtered dataset has only {} videos".format(len(coords)))
The original dataset has 6 videos
The filtered dataset has only 3 videos
We can also filter specific experiments with the .filter_videos() method, which takes a list of experiment IDs as input:
[21]:
single_video_coords = coords.filter_videos(['20191204_Day2_SI_JB08_Test_56'])
print("The new filtered dataset has only {} video".format(len(single_video_coords)))
The new filtered dataset has only 1 video
Last but not least, we can also keep all videos, but filter certain animals whose coordinates we’d like to keep for further analysis. As seen above, the dataset used in this tutorial contains two animals per video: a C57Bl6 (labelled “B”) and a CD1 (labelled “W”). Let’s see how we can keep only the C57B16 with the .filter_id() method:
Let’s first point out that, before filtering, a given experiment in the coords object has 44 features (22 from each animal).
[22]:
coords['20191204_Day2_SI_JB08_Test_56']
[22]:
| B_Center | B_Left_bhip | B_Left_ear | B_Left_fhip | B_Nose | ... | W_Right_ear | W_Right_fhip | W_Spine_1 | W_Spine_2 | W_Tail_base | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | x | y | x | y | x | y | x | y | ... | x | y | x | y | x | y | x | y | x | y | |
| 00:00:00 | 538.363647 | 249.284027 | 552.938171 | 238.256393 | 526.389221 | 298.484528 | 554.218140 | 263.846924 | 497.151367 | 300.893616 | ... | 163.311036 | 230.528197 | 151.194123 | 260.958313 | 145.208389 | 245.764724 | 123.380149 | 292.024324 | 117.470673 | 316.318786 |
| 00:00:00.039938658 | 538.369385 | 249.327469 | 552.927368 | 238.261719 | 526.412781 | 298.484711 | 554.220703 | 263.867645 | 497.139038 | 300.743500 | ... | 167.872041 | 221.538756 | 156.158676 | 252.000641 | 147.822707 | 238.749525 | 126.308410 | 282.404055 | 117.764466 | 305.007018 |
| 00:00:00.079877316 | 538.341919 | 249.331680 | 552.979980 | 238.308563 | 526.437988 | 298.471222 | 554.231751 | 263.856079 | 497.157898 | 300.888397 | ... | 172.082931 | 213.227768 | 160.587585 | 244.264328 | 151.865173 | 229.841355 | 129.889419 | 272.855470 | 119.838295 | 295.199738 |
| 00:00:00.119815975 | 538.328064 | 249.476318 | 553.003174 | 238.238953 | 526.427002 | 298.530182 | 554.242981 | 263.896118 | 497.196014 | 300.982727 | ... | 176.770797 | 203.171585 | 164.369293 | 235.348343 | 156.897583 | 219.281571 | 134.908768 | 261.188996 | 124.537338 | 280.778840 |
| 00:00:00.159754633 | 538.361267 | 249.156189 | 552.935913 | 238.178787 | 526.232361 | 298.454224 | 554.304565 | 263.707153 | 497.193512 | 300.888733 | ... | 184.708252 | 193.616104 | 168.269272 | 226.879074 | 161.464935 | 209.005005 | 135.903274 | 252.997026 | 125.637749 | 273.162354 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800306707 | 417.213013 | 120.369354 | 434.364075 | 121.799324 | 407.657898 | 149.051193 | 420.671143 | 132.926590 | 403.773225 | 168.571396 | ... | 458.965545 | 138.696243 | 478.439513 | 152.962585 | 464.861633 | 161.004669 | 492.194305 | 185.569657 | 507.282135 | 198.650726 |
| 00:09:58.840245366 | 418.764465 | 121.211830 | 437.265686 | 124.925872 | 408.706879 | 148.632080 | 423.572205 | 135.010574 | 403.317353 | 167.953598 | ... | 459.109069 | 139.845016 | 480.203215 | 153.216278 | 465.769653 | 161.612426 | 494.342407 | 186.055055 | 509.487884 | 199.299560 |
| 00:09:58.880184024 | 417.745423 | 119.924720 | 436.591827 | 123.725265 | 407.754945 | 145.980683 | 422.490540 | 133.576019 | 404.400087 | 162.526779 | ... | 459.507567 | 142.085800 | 482.209348 | 155.651763 | 468.152923 | 164.586746 | 496.519652 | 187.808962 | 510.491302 | 200.315918 |
| 00:09:58.920122683 | 417.745423 | 119.924720 | 436.591827 | 123.725265 | 407.754945 | 145.980683 | 422.490540 | 133.576019 | 404.400087 | 162.526779 | ... | 459.507567 | 142.085800 | 482.209348 | 155.651763 | 468.152923 | 164.586746 | 496.519652 | 187.808962 | 510.491302 | 200.315918 |
| 00:09:58.960061341 | 417.745423 | 119.924720 | 436.591827 | 123.725265 | 407.754945 | 145.980683 | 422.490540 | 133.576019 | 404.400087 | 162.526779 | ... | 459.507567 | 142.085800 | 482.209348 | 155.651763 | 468.152923 | 164.586746 | 496.519652 | 187.808962 | 510.491302 | 200.315918 |
14998 rows × 44 columns
After filtering to keep only the C57Bl6 (“B”), there are only 22 features left (scroll right to see that, as expected, no features related to “W” remain):
[23]:
coords.filter_id("B")['20191204_Day2_SI_JB08_Test_56']
[23]:
| B_Center | B_Left_bhip | B_Left_ear | B_Left_fhip | B_Nose | ... | B_Right_ear | B_Right_fhip | B_Spine_1 | B_Spine_2 | B_Tail_base | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | x | y | x | y | x | y | x | y | ... | x | y | x | y | x | y | x | y | x | y | |
| 00:00:00 | 538.363647 | 249.284027 | 552.938171 | 238.256393 | 526.389221 | 298.484528 | 554.218140 | 263.846924 | 497.151367 | 300.893616 | ... | 515.534424 | 278.740295 | 524.835266 | 258.351959 | 536.314819 | 269.603027 | 536.819580 | 230.367477 | 530.644226 | 211.947082 |
| 00:00:00.039938658 | 538.369385 | 249.327469 | 552.927368 | 238.261719 | 526.412781 | 298.484711 | 554.220703 | 263.867645 | 497.139038 | 300.743500 | ... | 515.557556 | 278.727722 | 524.861694 | 258.385285 | 536.332397 | 269.632355 | 536.820496 | 230.371612 | 530.645569 | 211.948883 |
| 00:00:00.079877316 | 538.341919 | 249.331680 | 552.979980 | 238.308563 | 526.437988 | 298.471222 | 554.231751 | 263.856079 | 497.157898 | 300.888397 | ... | 515.525696 | 278.735718 | 524.850891 | 258.373383 | 536.320923 | 269.619812 | 536.835327 | 230.389618 | 530.657898 | 211.960388 |
| 00:00:00.119815975 | 538.328064 | 249.476318 | 553.003174 | 238.238953 | 526.427002 | 298.530182 | 554.242981 | 263.896118 | 497.196014 | 300.982727 | ... | 515.538391 | 278.738708 | 524.852112 | 258.445496 | 536.338318 | 269.629272 | 536.825806 | 230.358459 | 530.663635 | 211.948959 |
| 00:00:00.159754633 | 538.361267 | 249.156189 | 552.935913 | 238.178787 | 526.232361 | 298.454224 | 554.304565 | 263.707153 | 497.193512 | 300.888733 | ... | 515.538025 | 278.705048 | 524.844055 | 258.242065 | 536.360901 | 269.499512 | 536.811890 | 230.327743 | 530.650696 | 211.961029 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 00:09:58.800306707 | 417.213013 | 120.369354 | 434.364075 | 121.799324 | 407.657898 | 149.051193 | 420.671143 | 132.926590 | 403.773225 | 168.571396 | ... | 389.377777 | 147.743684 | 402.071594 | 119.623657 | 406.150024 | 129.519211 | 430.530731 | 113.965172 | 448.407562 | 110.662156 |
| 00:09:58.840245366 | 418.764465 | 121.211830 | 437.265686 | 124.925872 | 408.706879 | 148.632080 | 423.572205 | 135.010574 | 403.317353 | 167.953598 | ... | 389.137940 | 146.219651 | 402.076691 | 119.482201 | 407.196777 | 129.546539 | 432.573608 | 114.848869 | 448.386139 | 111.038552 |
| 00:09:58.880184024 | 417.745423 | 119.924720 | 436.591827 | 123.725265 | 407.754945 | 145.980683 | 422.490540 | 133.576019 | 404.400087 | 162.526779 | ... | 387.196167 | 144.977769 | 399.367402 | 117.643234 | 406.196411 | 127.711792 | 431.796935 | 114.095085 | 448.092437 | 110.713548 |
| 00:09:58.920122683 | 417.745423 | 119.924720 | 436.591827 | 123.725265 | 407.754945 | 145.980683 | 422.490540 | 133.576019 | 404.400087 | 162.526779 | ... | 387.196167 | 144.977769 | 399.367402 | 117.643234 | 406.196411 | 127.711792 | 431.796935 | 114.095085 | 448.092437 | 110.713548 |
| 00:09:58.960061341 | 417.745423 | 119.924720 | 436.591827 | 123.725265 | 407.754945 | 145.980683 | 422.490540 | 133.576019 | 404.400087 | 162.526779 | ... | 387.196167 | 144.977769 | 399.367402 | 117.643234 | 406.196411 | 127.711792 | 431.796935 | 114.095085 | 448.092437 | 110.713548 |
14998 rows × 22 columns
Now that we have a basic understanding of how to create and interact with a project, coordinates, and table dictionaries, let’s show some plots!
Basic visual exploration
Let’s first see some basic heatmaps per condition. All plotting functions within DeepOF are hosted in the deepof.visuals module. Among many other things, we can plot average heatmaps per experimental condition! Let’s see if we can visualize ant interesting patterns on the available data:
[24]:
sns.set_context("notebook")
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
deepof.visuals.plot_heatmaps(
my_deepof_project,
["B_Nose"],
center="arena",
exp_condition="CSDS",
condition_value="Nonstressed",
ax=ax1,
show=False,
display_arena=True,
experiment_id="average",
)
deepof.visuals.plot_heatmaps(
my_deepof_project,
["B_Nose"],
center="arena",
exp_condition="CSDS",
condition_value="Stressed",
ax=ax2,
show=False,
display_arena=True,
experiment_id="average",
)
plt.tight_layout()
plt.show()
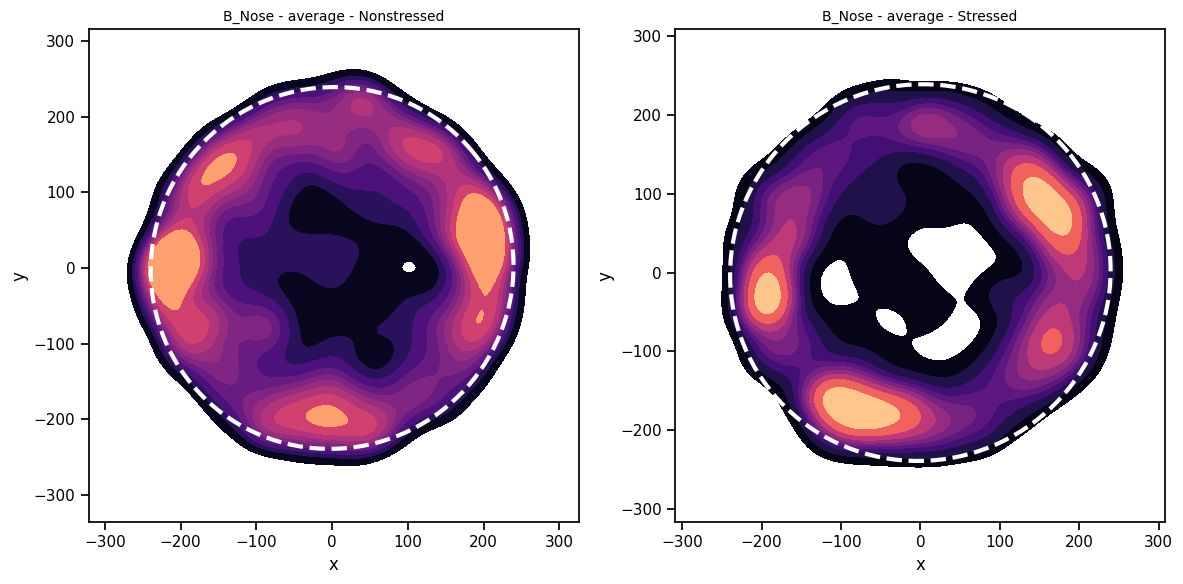
It seems stressed animals spend more time closer to the walls of the arena, and less time in the center! For details on how deepof.visuals.plot_heatmap() works, feel free to check the full API reference or the function docstring.
Finally, let’s create an animated video showing our newly preprocessed data. DeepOF can produce reconstructions of the tracks and show them as videos. All animals and the arena are displayed by default. This is particularly useful when interpreting clusters and visualizing embeddings in the unsupervised pipeline, as we’ll see in a later turorial.
[25]:
from IPython import display
video = deepof.visuals.animate_skeleton(
my_deepof_project,
experiment_id="20191204_Day2_SI_JB08_Test_54",
frame_limit=500,
dpi=60,
)
html = display.HTML(video)
display.display(html)
plt.close()
What’s next
That’s it for this tutorial. Next, we’ll see how to run a supervised annotation pipeline with pretrained models!